ML367 : ATAD5 (ATPase family AAA domain-containing protein 5) Inhibitor
ML367
Target Name
ATPase family AAA domain-containing protein 5
Target Alias
ATAD5
Target Class
DNA Metabolism Protein
Mechanism of Action
Inhibitor of ATAD5
Biological / Disease Relevance
DNA damage response, Cancer Biology
In vitro activity
ATAD5 bioassay (IC50)Cellular activity
HEK293 Cell viability assay (IC50)In vitro activity
Mouse Plasma Stability (T1/2)In vitro activity
PAMPA (10^-6 cm/s)Target Information
Encoding the genetic instructions essential to both our development and function as living organisms, our DNA must be maintained with exquisite precision and integrity, especially throughout replication (Ciccia 2010, Zhou 2007). DNA can undergo damage in many different ways by both endogenous and exogenous agents. Thus, the numerous mechanisms by which DNA damage is both recognized and repaired are essential to cell survival. ATAD5 is involved in the DNA damage response, and its protein level increases in response to DNA damage without an increase in mRNA transcription (Michod 2007, Lee 2013). Identification of pathway(s) that stabilize ATAD5 protein levels in response to DNA damage and inhibitors of these pathway(s) would be beneficial to understanding a novel mechanism involved in the DNA damage response and introduce a new therapeutic approach for sensitizing cancer cells, respectively (Bell 2011, Michod 2007). However, no chemical matter is currently known that perturbs ATAD5 function. To understand the biology of ATAD5 and to evaluate its therapeutic potential, we conducted a quantitative high throughput screening campaign and subsequent medicinal chemistry optimization in pursuit of small molecules that destabilize ATAD5. Herein, we detail the discovery of ML367, a probe molecule that has low micromolar inhibitory activity in the ATAD5 destabilizer screen run with 10 μM 5-fluorouridine (5-FUrd) as the DNA damaging agent. Interestingly, ML367 was found to block general DNA damage responses including RPA32-phosphorylation and CHK1-phosphorylation in response to UV irradiation. In this regard, the probe molecule could block DNA repair pathways that function upstream of ATAD5. Additionally, the compound sensitized cells possessing a knock-out mutation of the PARP1 gene and as a result may serve as a sensitizer to kill cancer cells defective in the poly (ADP-ribose) polymerase 1 (PARP1)-dependent DNA repair pathway (Rohde 2010).
Properties

ML367
NCGC00262816
| Physical & chemical properties | ||||
|---|---|---|---|---|
| Molecular Weight | 334.3 g/mol | |||
| Molecular Formula | C19H12F2N4 | |||
| cLogP | 4.2 | |||
| PSA | 50.7 Ų | |||
| Storage | ||||
| Solubility | ||||
| CAS Number | 381168-77-0 | |||
SMILES:
FC1=C(C=C(C=C1)NC2=NC(C3=CC=NC=C3)=NC4=CC=CC=C24)F
InChI:
1S/C19H12F2N4/c20-15-6-5-13(11-16(15)21)23-19-14-3-1-2-4-17(14)24-18(25-19)12-7-9-22-10-8-12/h1-11H,(H,23,24,25)
InChIKey:
LBPYNNJXARHGAG-UHFFFAOYSA-N
Activity
Summary activity statement /
ATAD5 is a known suppressor of genomic instability and tumor formation in mice. ATAD5 protein levels increase in response to DNA damage and thus an inhibitor of ATAD5 stabilization, such as ML367 (SID 161004434; CID 921541), could sensitize cancer cells to DNA damaging agents. In this study, ML367 exhibited inhibition of ATAD5 stabilization in HEK293T cells as well as destabilization of the protein by western blot analysis. Moreover, our results demonstrate that treatment of cells deficient in DNA damage repair proteins (e.g. PARP1, Lig3, Lig4, FancM, FancG, and Rad54b) with ML367 results in significant growth inhibition in colony formation assays. The ML367 can therefore be used by investigators as a tool to further understand the role of ATAD5 in repair mechanism. Moreover, these data suggest a potential use of ML367 in combination with inhibitors of DNA repair proteins (e.g. PARP1) and/or cancer cells deficient in enzymes involved in the DNA repair response.
As no prior art for inhibitors of ATAD5 stabilization exist, ML367 represents an important tool for the scientific community to begin to understand the protein’s role in DNA repair as well as other biological modalities. First, ML367 can be used to dissect initial events in the DNA damage response. The molecular mechanism of action in which ML367 destabilizes TEL2 will unveil how DNA damage can activate TEL2 and its downstream targets. Many of these targets have been suggested to play important roles in tumorigenesis. Additionally, a number of genetic disorders are the result of mutations in these genes. For instance, ataxia telansiectasia (AT) is caused by a mutation in ATM, Seckel syndrome is caused by a hypomorphic mutation in ATR, and severe combined immunodeficiency is caused by a mutation in DNA-PKcs. As no prior art exists for inhibitors of ATAD5 or TEL2 stabilization, ML367 is positioned to be a novel tool to study the molecular mechanism of DNA damage response and the resultant signal cascade with a potentially wide application to cancer and other genetic diseases.
In vitro activity - Selectivity and Cytotoxicity Assay
| Bioassay | ML367 (IC50) |
|---|---|
|
ATAD5 |
1.2 uM |
|
CMV-Luc (Anti-Target) |
> 46 uM |
|
Luciferase Bioassay (Anti-Target) |
> 57 uM |
|
HEK293-viability |
Not toxic |
Summary /
The probe is found to have > 40 fold selective against the ATAD5 vs. CMV-Luc anti-target; and > 50 fold vs. luciferase biochemical assay. The compound probe also showed no apparent cytotoxicity.

Figure 1. Dose response curves for the probe ML367 against the ATAD5-Luc primary screen (green) and the cell viability assay (red). Results showed ML367’s dose response inhibition of ATAD5 activity without any significant cytotoxic effect.
Cellular activity - Nuclear Receptor Profiling Assay
Summary /
To evaluate the cellular activity of the probe, a secondary assay using a different cell line was developed. HEK293T cells were transfected with FLAG-tagged ATAD5 using Lipofectamine 2000 (Life Technologies), according to the manufacturer’s protocol. 48 hours post-transfection, the cells were treated with the indicated compounds for 16 hours. To obtain total lysate, the cells were resuspended in lysis buffer [50 mM Tris, pH 7.5, 150 mM NaCl, 1% Nonidet P-40, 5 mM EDTA, protease inhibitors (Roche)] and lysed on ice for 30 min. Proteins were separated by SDS-PAGE using a 4–15% Tris-glycine gel (Bio-Rad) and transferred to a Polyvinylidene difluoride membrane. FLAG-ATAD5 protein levels were detected by the ECL Western Blotting Detection System (GE Healthcare) using an HRP conjugated antibody against FLAG (Sigma). Equal protein loading was confirmed using an antibody against tubulin (Abcam). The ratio of FLAG/Tubulin was quantified using ImageJ.
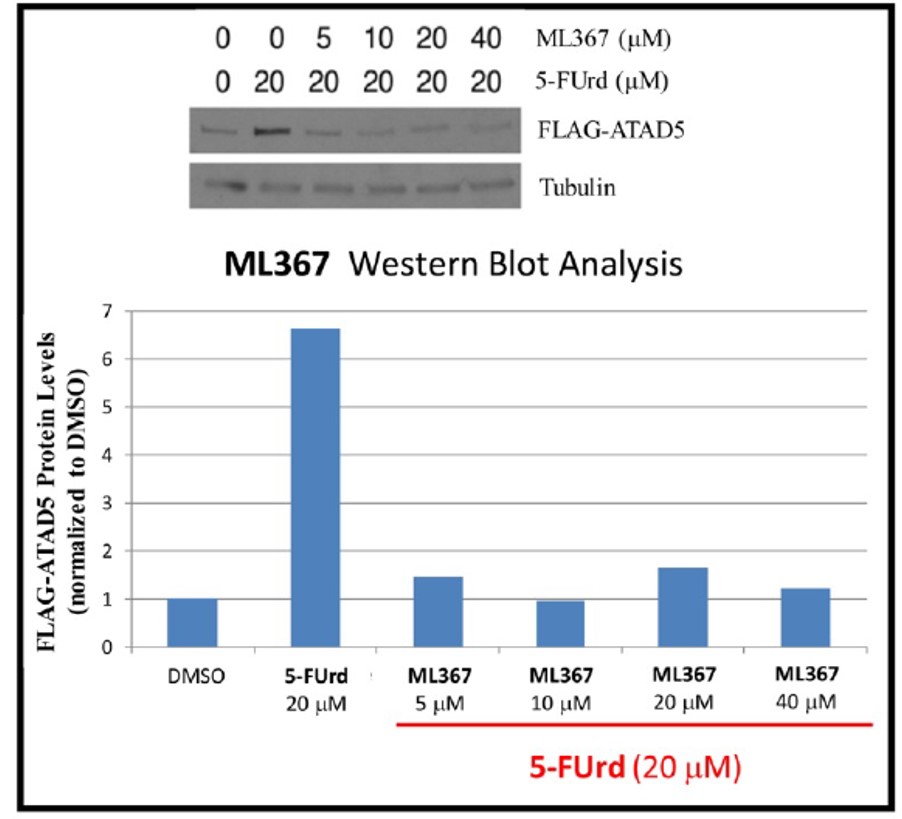
Figure 1. Inhibition of FLAG-ATAD5 stabilization by ML367 HEK293T cells were transiently transfected with FLAG-tagged ATAD5 and treated with the indicated amount of ML367 for 16 hours in the presence or absence of 20 μM 5-FUrd. ATAD5 protein levels were visualized by western blotting using an antibody against FLAG (top panel), and quantified using ImageJ (bottom panel). Results showed inhibition of ATAD5 expression upon treatment of the probe.
In vitro activity - ADME Profiling
Summary /
The preliminary absorption, distribution, metabolism, excretion (ADME) profile of ML367 supports its use as a valuable probe for ATAD5 destabilization. While both its microsomal stability (in rat and human) and solubility (in PBS buffer) are moderate, the latter was above the IC determined in the cell based assay. ML367 has good PAMPA permeability, and its overall ADME profile is consistent with its measured Log D of 1.58 (Table 1). Additionally, ML367 showed good stability in mouse plasma as well as a series of aqueous stability assessments including pH 2 and pH 10 buffers and aqueous 5 mM glutathione. While the overall ADME profile may limit the utility of ML367 in vivo, results of solubility, permeability and biological activity showed good results.
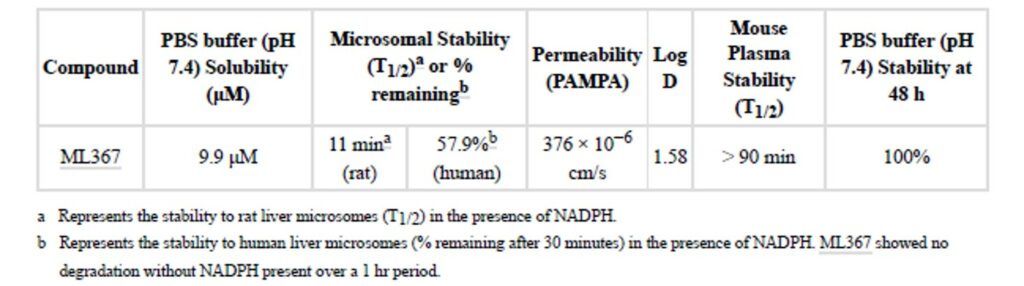
Table 1. ADME profile of ML367.
References
- qHTS assay for small molecules that inhibit ELG1-dependent DNA repair in human embryonic kidney (HEK293T) cells expressing luciferase-tagged ELG1: Summary
- Rohde JM, Rai G, Choi YJ, et al. Discovery of ML367, inhibitor of ATAD5 stabilization. 2013 Apr 15 [Updated 2013 Nov 14]. In: Probe Reports from the NIH Molecular Libraries Program [Internet]. Bethesda (MD): National Center for Biotechnology Information (US); 2010
- Ciccia A, Elledge SJ. The DNA damage response: making it safe to play with knives. Mol Cell. 2010;40(2):179-204. doi:10.1016/j.molcel.2010.09.019
- Zhou T, Chou J, Zhou Y, et al. Ataxia telangiectasia-mutated dependent DNA damage checkpoint functions regulate gene expression in human fibroblasts. Mol Cancer Res. 2007;5(8):813-822. doi:10.1158/1541-7786.MCR-07-0104
- Michod D, Widmann C. DNA-damage sensitizers: potential new therapeutical tools to improve chemotherapy. Crit Rev Oncol Hematol. 2007;63(2):160-171. doi:10.1016/j.critrevonc.2007.04.003
- Lee KY, Fu H, Aladjem MI, Myung K. ATAD5 regulates the lifespan of DNA replication factories by modulating PCNA level on the chromatin. J Cell Biol. 2013;200(1):31-44. doi:10.1083/jcb.201206084
- Bell DW, Sikdar N, Lee KY, et al. Predisposition to cancer caused by genetic and functional defects of mammalian Atad5. PLoS Genet. 2011;7(8):e1002245. doi:10.1371/journal.pgen.1002245


