ML265 : PKM (Pyruvate Kinase M2) Activator
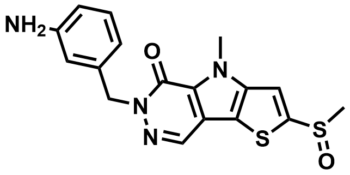
ML265
Target Name
Pyruvate Kinase M2
Target Alias
PKM
Target Class
Kinase
Mechanism of Action
Activator of PKM
Biological / Disease Relevance
Tumor cell growth and proliferation, Glucose metabolism
In vitro activity
hPyk-M2 bioassay (IC50)In vitro activity
LDh secondary assay (IC50)In vivo activity
Mouse xenograft (tumor size)Target Information
Cancer cells have altered metabolic processes compared to normal differentiated cells and the expression of the M2 isozyme of pyruvate kinase (PKM2) plays an important role in this aberrant metabolism. The M1 isoform is a highly active enzyme typically expressed in muscle and brain tissue, the alternatively spliced M2 variant is considerably less active and expressed in many tumors studied to date. This report describes the use of the PKM2 activator, ML265, and details some of the biophysical, ex vivo and in vivo activity of this compound. ML265 induces the more active tetrameric state of PKM2 and the X-ray co-crystal structure shows that the activator binds at the dimer-dimer interface between two subunits of PKM2. This compound was tested in a H1299 mouse xenograft model and showed significant reduction in tumor size, weight, and occurrence with no apparent toxicity over the 7-week experiment.
Properties

ML265
TEPP-46
| Physical & chemical properties | ||||
|---|---|---|---|---|
| Molecular Weight | 372.5 g/mol | |||
| Molecular Formula | C17H16N4O2S2 | |||
| cLogP | 1.3 | |||
| PSA | 128 Ų | |||
| Storage | ||||
| Solubility | ||||
| CAS Number | 1221186-53-3 | |||
SMILES:
CN1C2=C(C(C=NN3CC4=CC(N)=CC=C4)=C1C3=O)SC(S(C)=O)=C2
InChI:
1S/C17H16N4O2S2/c1-20-13-7-14(25(2)23)24-16(13)12-8-19-21(17(22)15(12)20)9-10-4-3-5-11(18)6-10/h3-8H,9,18H2,1-2H3
InChIKey:
ZWKJWVSEDISQIS-UHFFFAOYSA-N
Activity
Summary activity statement /
ML265 (SID 85267885; CID 44246499) is a potent activator of PKM2 in both biochemical (AC50 = 92 nM) and cell-based assays with high selectivity over PKM1, PKR and PKL. The compound was found to induce tetramerization based upon size exclusion chromatography, sucrose gradient ultracentrifugation and immunoprecipitation experiments. A high resolution X-ray structure revealed that ML265 binds at the dimer-dimer interface of the PKM2 homotetramer. Importantly, this compound is capable of activating PKM2 in cell lysate of pervanadate treated cells, which is a condition known to inhibit PKM2 activity through accumulation of phosphotyrosine peptides. ML265 significantly increased the doubling time of H1299 cells under hypoxic conditions, but interestingly showed no effect under normoxia. PKM2 expression has also been shown to effect PGAM1 phosphorylation and ML265 was able to reduce this phosphorylation event. After we determined that ML265 had an acceptable in vitro microsomal stability profile, it was advanced into in vivo PK experiments and ultimately showed a reduction in tumor size, weight, and occurrence in a H1299 mouse xenograft model.
In vitro activity - Selectivity and Cytotoxicity Assay
| Bioassay | ML265 (IC50) |
|---|---|
|
hPyk-M2 |
92 nM |
|
hPyk-M1 (Anti-Target) |
Inactive |
|
hPyk-R (Anti-Target) |
Inactive |
|
hPyk-L (Anti-target) |
Inactive |
Summary /
The tetrameric state is the most active form of pyruvate kinase, which is induced by the endogenous activator 1,6-FBP for PKL, PKR and PKM2. Typically, this feed forward mechanism allows cells to respond to high glycolytic flux and process the resulting PEP to pyruvate and ATP in an appreciably exothermic reaction (ΔGo’ = -7.5 kcal/mol). Cancer cells express PKM2 and somewhat paradoxically exhibit high rates of aerobic glycolysis while having remarkably low pyruvate kinase activity when compared to PKM1 expressing cells. In line with our previously reported activators, ML265 potently activates PKM2 in vitro with an AC50 = 92 nM and shows a high degree of selectivity over the other 3 pyruvate kinase isoforms (Figure 1).
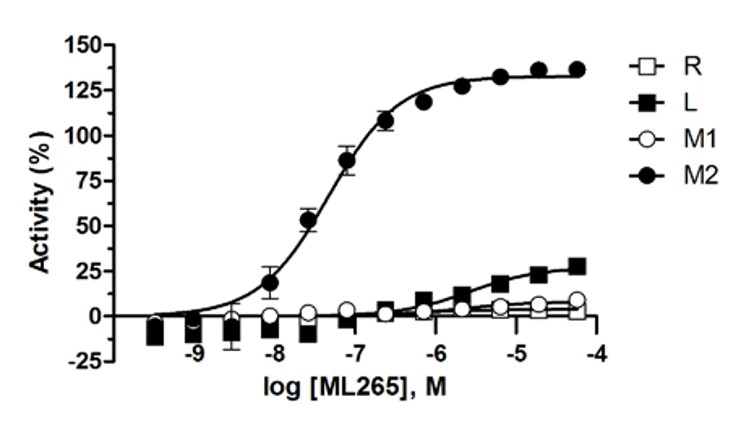
Figure 1. ML265 shows selective activation of PKM2 over PKM1, PKR and PKL in the luminescent pyruvate kinase activity assays using KinaseGlo.
Cellular activity - Functional assays
Summary /
Cancer cells are known to exist in highly phosphorylated states and certain peptide motifs with phosphorylated tyrosines are known PKM2 inhibitors (1Christofk 2008, Ikeda 2000). Binding of these phosphorylated peptides results in 1,6-FBP extrusion from the allosteric site. Use of a known phosphorylated peptide motif that binds to PKM2 (pM2tide) causes a shift in abundance of the more active tetrameric state to the less active monomeric state compared to the same, non-phosphorylated peptide (M2tide) (Figure 2a). ML265 was able to maintain potent activation of PKM2 in the presence of both M2tide and pM2tide (Figure 2b). Treating cells with pervanadate, a known phosphatase inhibitor, causes increased amounts of phosphorylated proteins and decreased PKM2 activity (1Christofk 2008). To test whether ML265 could activate PKM2 in this cellular context, we determined pyruvate kinase activity in cell lysate of both pervanadate and non-pervanadate treated cells (Figure 2c). Remarkably, at 1 μM, ML265 was able to activate PKM2 regardless of pervanadate treatment. Cantley and co-workers have shown that RNAi mediated replacement of PKM2 with PKM1 results in higher pyruvate kinase activity and reduces cellular proliferation in a hypoxic environment (2Christofk 2008).
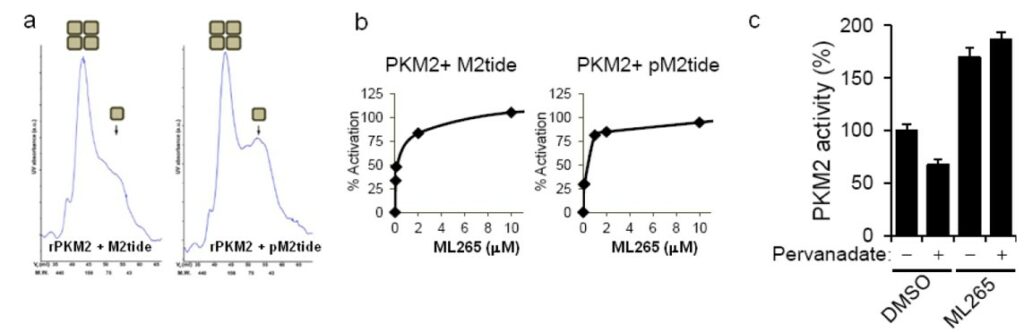
Figure 2. Figure 4. ML265 activates PKM2 in the presence of phosphorylated peptides. a) Size exclusion chromatography of recombinant PKM2 after incubation with an M2 binding peptide motif (M2tide) or phosphorylated version of the same peptide motif (pM2tide). b) ML265’s ability to activate PKM2 in the presence of M2tide or pM2tide. c) Pyruvate kinase activity in A549 cell lysates following treatment with or without pervanadate in the presence of either DMSO or ML265.
In vitro activity - Mechanism of action
Summary /
ML265 activates PKM2 via an increase in PEP affinity (Jiang 2010). In the absence of activator, PKM2 has a relatively low affinity for PEP (KM ~ 1.5 mM), but treatment with either 1,6-FBP or ML265 resulted in a significant shift in KM to ~0.08 and ~0.09 mM respectively (Figure 3a). In accordance with our previous studies, this effect was not seen with respect to ADP affinity (Figure 3b). The homotetramer of PKM2 is the most active form and its endogenous activator, 1,6-FBP is thought to induce this subunit association. To investigate this hypothesis, we used size exclusion chromatography to separate the monomers from tetramers and then determined the effect of 1,6-FBP on PKM2 activity (Figure 4a). As expected, the untreated tetramer had considerably higher kinase activity than the untreated monomer. Addition of 100 μM 1,6-FBP had little effect on the tetramer activity, but resulted in ~70% increase in activity for the monomer (Figure 4b). In an effort to understand whether our activators could induce the more active tetrameric state, we incubated ML265 with recombinant PKM2 and then used sucrose gradient ultra-centrifugation followed by SDS-PAGE to separate the protein. As can be seen in Figure 4c, ML265 caused a significant shift to tetrameric species, similar to PKM1 and consistent with the affect seen with 1,6-FBP on PKM2.
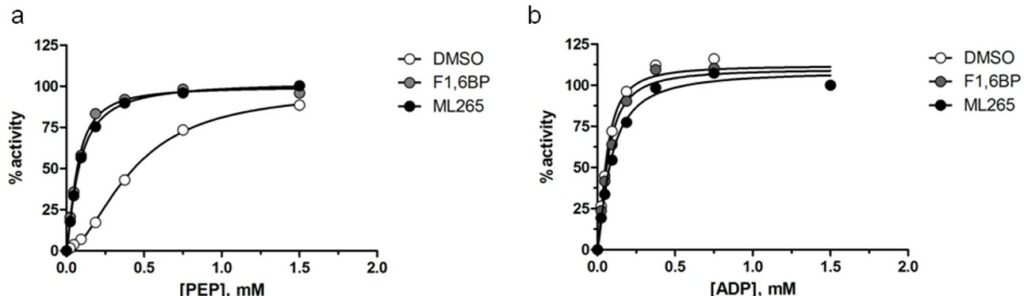
Figure 3. PKM2 activation (scaled from initial velocities) as a function of a) PEP and b) ADP concentration in the presence of 10μM 1,6-FBP (gray circles), ML265 (black circles) or DMSO control (white circles).
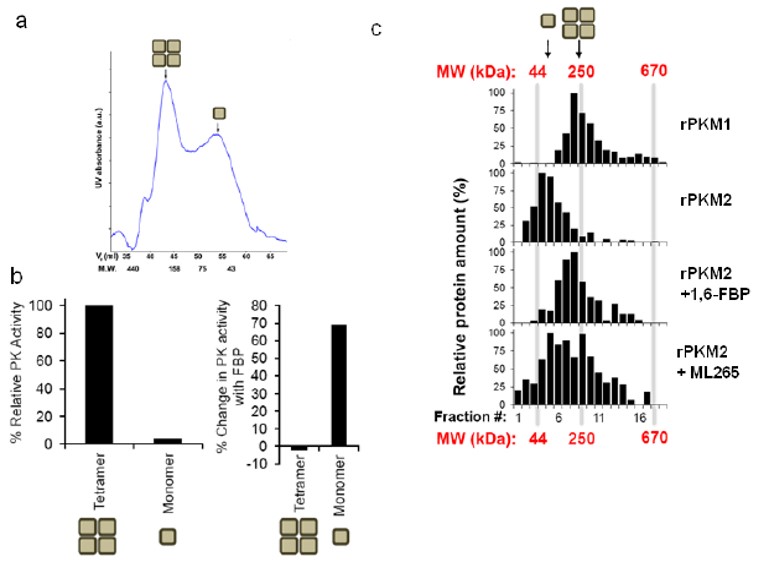
Figure 4. a) Size exclusion chromatography was used to separate monomeric and tetrameric PKM2 species and the fractions were b) assayed for pyruvate kinase activity with and without 1,6-FBP (100 μM). c) Sucrose gradient ultracentrifugation profiles of recombinant PKM1 or PKM2 and effects of 1,6-FBP or ML265 on PKM2 subunit stoichiometry.
In vitro activity - X-ray Crystallography
Summary /
A high resolution X-ray structure of ML265 bound to PKM2 (Figure 5) is generated. The structure showed that two activators and four 1,6-FBP molecules bind per tetramer. The two equivalents of ML265 bind at the dimer-dimer interface and are completely buried within this interfacial pocket. ML265 is accommodated through van der Waals interactions and water mediated hydrogen bonds to the pocket lining residues (Figure 5b).
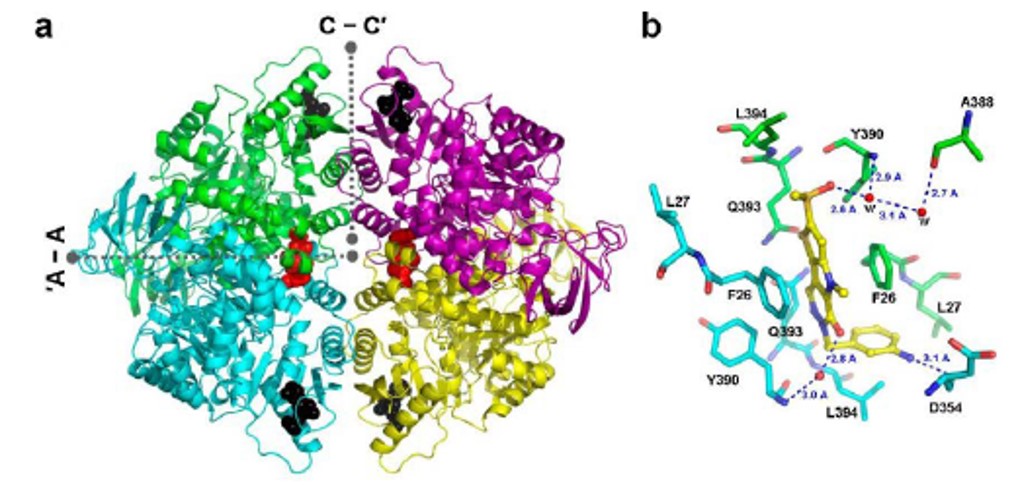
Figure 5. a) X-ray structure of the PKM2 tetramer with two equivalents of ML265 and four equivalents of 1,6-FBP bound. ML265 binds at the dimer-dimer interface, termed A-A’. b) The sulfoxide and carbonyl group of ML265 form water mediated hydrogen bond networks the protein, while the aniline moiety forms a direct hydrogen bond with D354.
In vitro and vivo activity - ADME and PK profiling
Summary /
ML265 gave superior plasma concentrations (vs the prior art PKM2 activator ML202) that persisted at higher levels over the 24 hour study (Figure 6). ML265 also displayed good oral bioavailability, low clearance, a long half-life and good volume of distrubtion (Table 1). These characteristics were deemed appropriate for use in the mouse xenograft efficacy model.
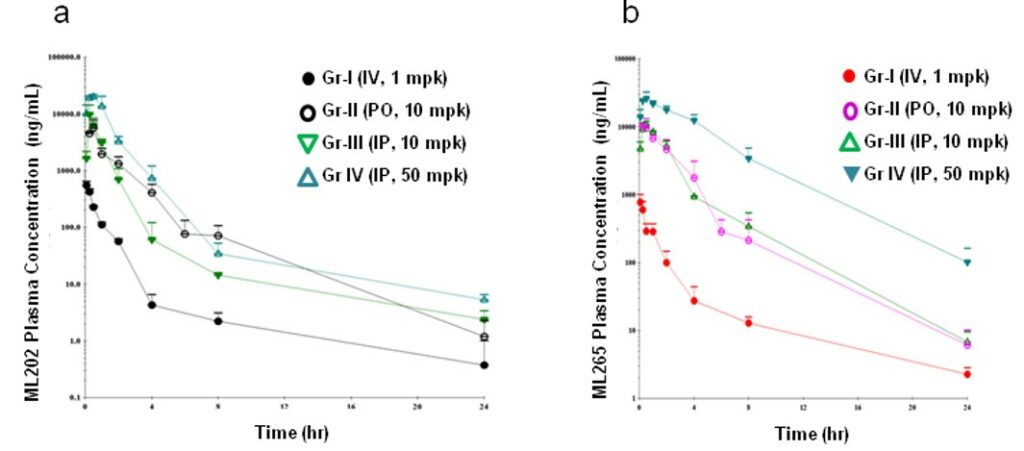
Figure 6. Plasma concentration-time profiles of a) ML202 and b) ML265 in male Balb/c mice following intravenous, intraperitoneal and oral administration.
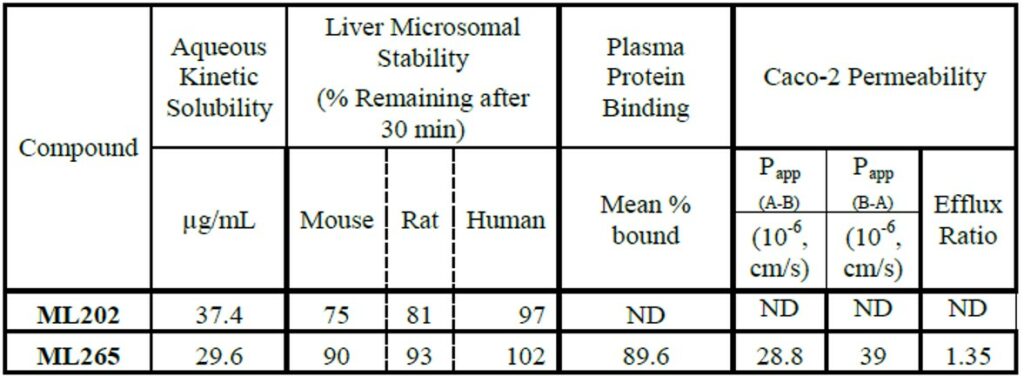
Table 1. In vitro ADME profile for ML202 and ML265.
In vivo activity - Functional assay: Mouse xenograft study
Summary /
ML265 was advanced into a 7-week H1299 mouse xenograft study using immunocompromised (nu/nu) mice. The mice were randomly divided into two cohorts with one group receiving the established 50 mg/kg BID of ML265, the other cohort received only vehicle. Blood counts, serum chemistries and histological examination of various tissues were followed to determine if there were overt toxicities associated with ML265 treatment (Table 2). No significant aberrations were observed in these toxicity measures when compared to the vehicle treated mice. ML265 was able to delay tumor formation compared to vehicle-treated mice and the tumors that did form were significantly smaller (Figure 7). Further examination of the tumors revealed that ML265 was detectable signifying that the tumor was being exposed to the activator.

Table 2. Serum chemistries and blood counts of mice treated with ML265. *WBC: White Blood Cells. §PCV: Packed Cell Volume.
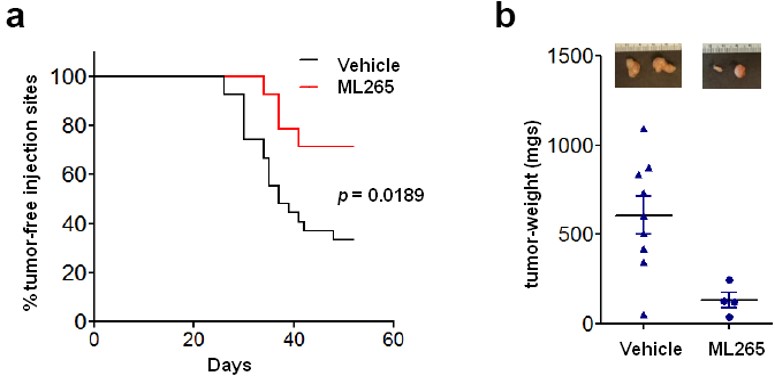
Figure 7. H1299 cells were injected subcutaneously into nu/nu mice which were subsequently randomly divided into two cohorts, one given vehicle and the other 50 mg/kg BID ML265 throughout the duration of the experiment. a) Injection sites were monitored for tumor emergence [p value calculated by log rank (Mantel-Cox) test]. b) At 52 days, the tumors were dissected and final tumor weights were measured. Mean tumor weights ± s.e.m. are shown and p value was calculated by unpaired Student’s t-test.
References
- PubChem link: Extended Characterization of Activators of Human Muscle isoform 2 Pyruvate Kinase: Summary
- Jiang J, Walsh MJ, Brimacombe KR, et al. ML265: A potent PKM2 activator induces tetramerization and reduces tumor formation and size in a mouse xenograft model. 2012 Mar 16 [Updated 2013 May 8]. In: Probe Reports from the NIH Molecular Libraries Program [Internet]. Bethesda (MD): National Center for Biotechnology Information (US); 2010-. Available from: https://www.ncbi.nlm.nih.gov/books/NBK153222/
- 1Christofk HR, Vander Heiden MG, Wu N, Asara JM, Cantley LC. Pyruvate kinase M2 is a phosphotyrosine-binding protein. Nature. 2008;452(7184):181-186. doi:10.1038/nature06667
- Ikeda, Y., Taniguchi, N. & Noguchi, T. Dominant negative role of the glutamic acid residue conserved in the pyruvate kinase M(1) isozyme in the heterotropic allosteric effect involving fructose-1,6-bisphosphate. J Biol Chem 275, 9150-6 (2000).
- 2Christofk HR, Vander Heiden MG, Harris MH, et al. The M2 splice isoform of pyruvate kinase is important for cancer metabolism and tumour growth. Nature. 2008;452(7184):230-233. doi:10.1038/nature06734
- Jiang JK, Boxer MB, Vander Heiden MG, et al. Evaluation of thieno[3,2-b]pyrrole[3,2-d]pyridazinones as activators of the tumor cell specific M2 isoform of pyruvate kinase. Bioorg Med Chem Lett. 2010;20(11):3387-3393. doi:10.1016/j.bmcl.2010.04.015


