ML199 : APEX1 (DNA-(apurinic or apyrimidinic site) endonuclease) Inhibitor
ML199
Target Name
DNA-(apurinic or apyrimidinic site) endonuclease
Target Alias
APEX1
Target Class
Carbon-oxygen Lyase
Mechanism of Action
Inhibitor of APEX1
Biological / Disease Relevance
Base Excision Repair (BER) Pathway, DNA damage
In vitro activity
APE1 bioassay (IC50)Cellular activity
RIA gel assay (IC50)Target Information
Probe compound, ML199 (CID 46925884, SID 99430950), and related inhibitors of Apurinic/apyrimidinic (AP) endonuclease (APE1) are reported herein. APE1 is a key component of the base excision repair (BER) pathway that is responsible for repair of DNA damage caused by many anti-cancer agents such as bleomycin and temozolomide. As a result, inhibition of APE1 has been postulated as a viable strategy for sensitizing tumor cells to chemotherapy. ML199 and its related analogs belong to a drug-like series that was identified and optimized through a focused medicinal chemistry effort to afford compounds which display competitive inhibition of APE1 activity in the low micromolar potency range. On target effect of the ML199 was demonstrated through a concentration depended inhibition of AP site incision activity in whole cell HeLa extracts. Moreover, ML199 potentiated the cytotoxicity of the DNA alkylating agent methylmethane sulfonate (MMS) at non-cytotoxic doses of the probe compound. The probe and its general class of compounds have shown to have good kinetic solubility, Caco-2 permeability, metabolic stability and other favorable physicochemical properties, thus making ML199 an ideal starting point for further pre-clinical development of anti-cancer agents.
Properties

ML199
NCGC00185090
| Physical & chemical properties | ||||
|---|---|---|---|---|
| Molecular Weight | 315.4 g/mol | |||
| Molecular Formula | C15H13N3OS2 | |||
| cLogP | 2.2 | |||
| PSA | 111 Ų | |||
| Storage | ||||
| Solubility | PBS buffer (pH 7.4 @ 25 C): 51.6 μM | |||
| CAS Number | ||||
SMILES:
CC(NC1=C(C2=C(S1)CNC2)C3=NC4=C(C=CC=C4)S3)=O
InChI:
1S/C15H13N3OS2/c1-8(19)17-14-13(9-6-16-7-12(9)21-14)15-18-10-4-2-3-5-11(10)20-15/h2-5,16H,6-7H2,1H3,(H,17,19)
InChIKey:
MODNMLYGANOZDZ-UHFFFAOYSA-N
Activity
Summary activity statement /
ML199 can be used for target validation of APE1 in enzymatic and cell-based systems. It can also be used in combination with known DNA cancer chemotherapeutics, such as temozolomide (TMZ), to potentiate the activity of these agents. It can be utilized in studies involving the BER pathway and the potentiation of known cancer chemotherapeutics. Lastly, the general class of this probe and its analogs has good in vitro ADME properties, making the probe series useful in further optimization for potential in vivo exposure.
In vitro activity - Selectivity Assay
| Bioassay | (IC50) |
|---|---|
|
APE1 |
6 uM |
|
Thiazole orange counterscreen (Anti-Target) |
>57 uM |
|
APE1 Radiotracer Incision Assay (RIA) |
1 uM |
Summary /
ML199 is found to have > 10 fold selective against the APE1.
In vitro activity - Secondary assay: Radiotracer Incision Assay (RIA)
Summary /
ML199 exhibited good potency in the secondary radiotracer assay (Figure 1) and gave a favorable ADME profile.
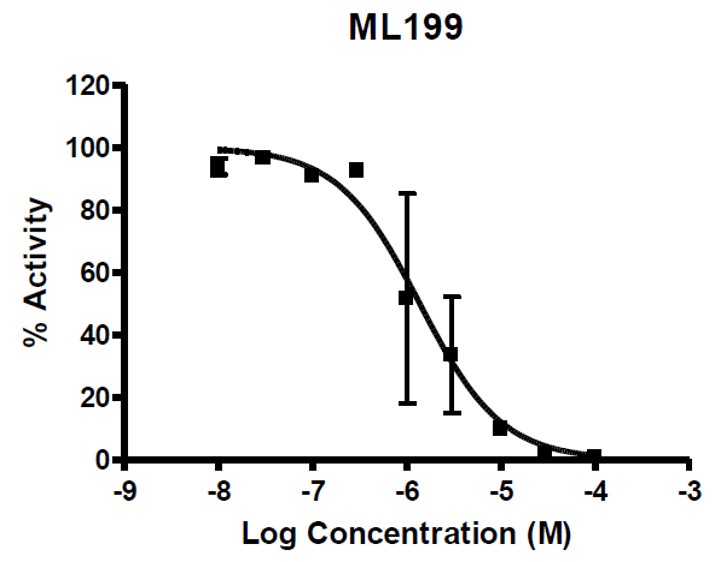
Figure 1. Dose response from ML199 in Radiotracer Incision Assay and structure of the probe. Results showed dose response inhibition upon treatment with ML199.
Cellular activity - Cytotoxicity Assessment: MTT and MMS Potentiation Assays
| Cell line | 5 uM | 20 uM | 50 uM |
|---|---|---|---|
|
HeLa (% viability) |
98 | 79 | ~5 |
|
T98G (% viability) |
99 | 95 | ~80 |
|
U-87 MG (% viability) |
35 | 20 | ~20 |
|
U-138 MG (% viability) |
92 | 80 | ~50 |
Summary /
ML199 was also shown to potentiate the toxicity of DNA-damaging agent MMS in a subsequent cell-based assay (Figure 2) while exhibiting only moderate cytotoxicity with the probe compound alone.
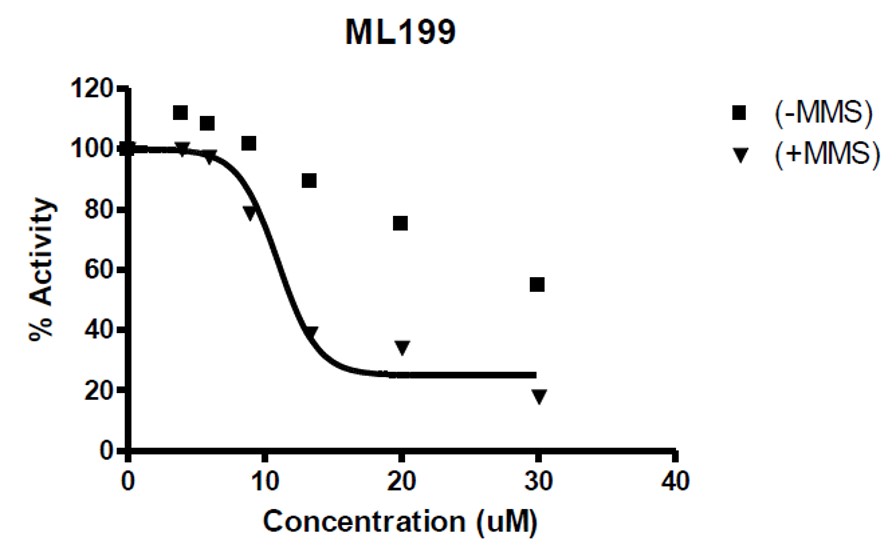
Figure 2. Potentiation of probe ML199 in the presence or absence of MMS.
Cellular Activity - Mechanism of Action
Summary /
Mechanism of action of the probe and the original hit were explored, and both compounds were shown to act as competitive inhibitors of APE1 (Figure 3). An electrophoretic mobility shift assay (EMSA), was employed to examine the stability of the APE1-DNA substrate complex in the presence of ML199 (Figure 4). The percent APE1-DNA complex decreased with the increasing concentration of the probe compound, further supporting the notion that ML199 acts by competitive inhibition and binds to Ape1 at the same site as the DNA substrate. On target effect of the probe was explored using HeLa whole cell extract incision assay in which ML199 was shown to inhibit AP Site incision activity which comparable activity as the purified enzyme assay (Figure 5).
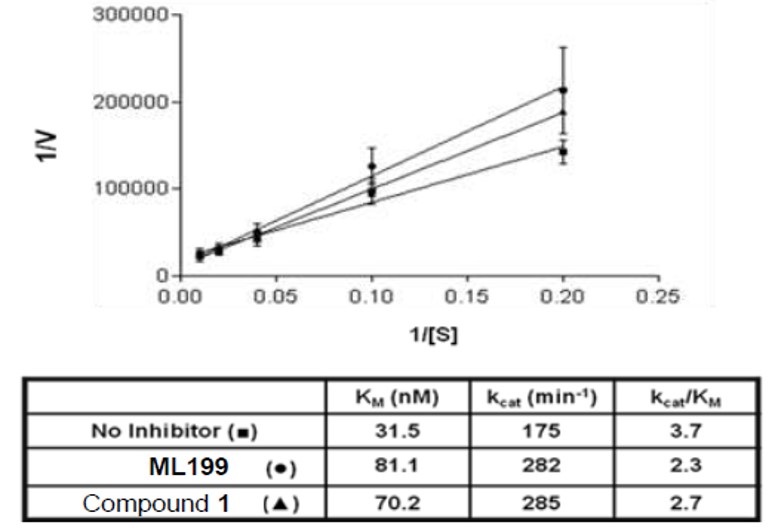
Figure 3. Kinetic parameters without and with ML199 or Compound 1. Ten pg of APE1 (~28 pM) was incubated without or with 5 μM of the indicated inhibitor at room temperature for 15 min. Increasing concentrations of radiolabeled abasic DNA substrate (i.e. 5, 10, 25, 50, or 100 nM) were then added, and the reactions were incubated at 37 ºC for 5 min before the addition of stop buffer. Intact substrate was separated from incised product on a 15% polyacrylamide denaturing gel, and the percent conversion was determined by standard phosphorimager analysis. Lineweaver – Burk plots of 1/V versus 1/[S] were used to determine KM and kcat (shown). The plotted data points (averages and standard deviations) were derived from 11 independent values for the no inhibitor reactions, and 5 values for each of the inhibitor points.
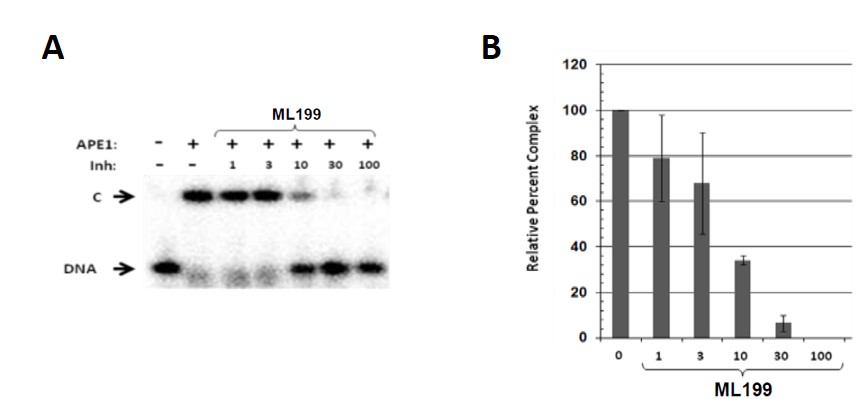
Figure 4. Stability of the APE1-DNA substrate complex in the presence of ML199. (A) Representative EMSA. Three hundred ng of APE1 (~0.8 μM) was incubated without inhibitor (final concentration 1% DMSO) or with the indicated inhibitor (1, 3, 10, 30 or 100 μM) for 10 min on ice. One hundred fmol of abasic DNA substrate (10 nM) was then added, and the binding reaction was incubated on ice for an additional 5 min. At that time, samples were subjected to non-denaturing polyacrylamide gel electrophoresis to separate the APE1-DNA complex (“C”) from unbound radiolabeled DNA (“DNA”). Inh = inhibitor. (B) Relative complex formation without (“0”) or with the indicated inhibitor (in μM). Shown is the average and standard deviation of three independent experimental data points, all relative to the APE1 control, without inhibitor.
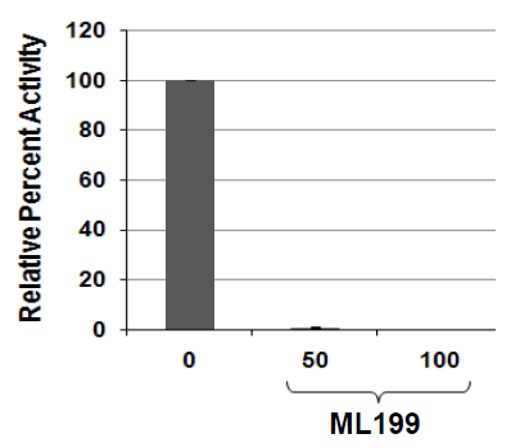
Figure 5. Inhibition of HeLa whole cell extract AP site incision activity with ML199. Three hundred ng of HeLa whole cell extract was incubated with 0, 50 or 100 uM of the indicated inhibitor at room temperature for 15 min, prior to the addition of 0.5 pmol radiolabeled AP-DNA substrate and subsequent transfer of the reaction mix (final volume of 10 μL) to 37 ºC for 5 min to allow for incision. Following addition of stop buffer and heat denaturation, the reaction products were subjected to 15% polyacrylamide denaturing gel electrophoresis. Shown is a bar graph reporting the relative percent incision activity in comparison to the no inhibitor control, arbitrarily set at 100. The values reported represent the averages and standard deviation of three independent experimental data points.
In vitro activity - ADME Profiling
Summary /
ML199 have many favorable attributes, yet a few liabilities exist. It is choosen as a probe molecule because of its greatly improved potency in the secondary radio-tracer assay (1 μM for ML199). The probe molecule also exhibits improved kinetic solubility, Caco-2 permeability and 10-fold less inhibition of the hERG channel or the HTS “hit” compound. Though the probe molecule is susceptible to mouse liver microsomes, compound 1 shows favorable stability. This seems to indicate that the general core scaffold is metabolically stable, and through additional structural modifications, this potential liability of the probe molecule could be addressed while maintaining potency.
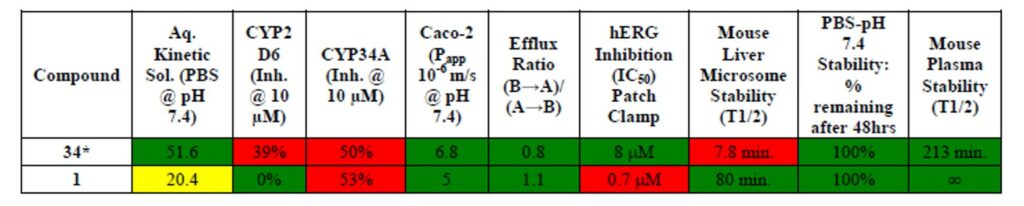
Table 1. In vitro ADME to date. Green = desirable property, Yellow = less than desirable property, Red = undesirable property. All experiments were conducted by Shanghai Chempartner Co. Ltd. 34* indicates probe molecule ML199.
References
- Probe Development Summary of Inhibitors of the Human Apurinic/apyrimidinic Endonuclease 1 (APE1)
- Rai G, Vyjayanti VN, Dorjsuren D, et al. Small Molecule Inhibitors of the Human Apurinic/apyrimidinic Endonuclease 1 (APE1). In: Probe Reports from the NIH Molecular Libraries Program. Bethesda (MD): National Center for Biotechnology Information (US); October 29, 2010


