ML196 : DYRK1A (Dual Specificity Tyrosine Phosphorylation Regulated Kinase 1A) Inhibitor
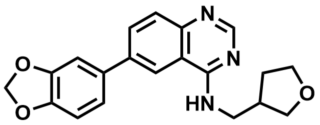
ML196
Target Name
Dual Specificity Tyrosine Phosphorylation Regulated Kinase 1A
Target Alias
DYRK1A
Target Class
Serine/ Threonine Protein Kinase
Mechanism of Action
Inhibitor of DYRK1A
Biological / Disease Relevance
Splicing modulator, pre-mRNA splicing, Gene splicing kinase
In vitro activity
Clk4 bioassay (IC50)In vitro activity
Clk1 bioassay (IC50)In vitro activity
Dyrk1A (IC50)In vitro activity
Clk4 assay (Kd)Target Information
The cdc2-like kinase (Clk) family contains four isoforms (Clk1-4) and is proposed to alter the function of the spliceosome by phosphorylating serine-arginine-rich (SR) proteins within the spliceosome assembly (Prasad 2003). The spliceosome regulates the processing, or splicing, of pre-mRNAs, yielding mature protein-encoding mRNAs (Black 2003, Wang 2008). Many human genes express more than one mRNA via alternative splicing, leading to protein diversity (Faustino 2003); however, misregulation of alternative splicing is involved in the pathogenesis of cancer and other diseases (He 2009, Matlin 2005). Studies have revealed that Clk isoforms are associated with alternative splicing of PkcβII (Jiang 2009), TF (Schwertz 2006), β-globin (Muraki 2004) and E1A pre-mRNA (Yomoda 2008). The Clks also regulate the alternative splicing of microtubule-associated protein tau and are implicated in frontotemporal dementia and Parkinson’s disease through the phosphorylation of splicing factors (SF) (Hartmann 2001). Inhibitors of Clk isoforms may alter these events and could prove to be useful agents in disease phenotypes characterized by abnormal splicing. Hagiwara reported TG003, a small molecule benzothiazole, as having low-nanomolar IC50 values versus Clk1 and Clk4. The patent literature revealed structurally similar benzothiazole 1a from Sirtris Pharmaceuticals (Perni 2009) and a quinoline 3 from Chronogen, Inc. reported to have activity versus Clk1 (Figure 1). Indole 2 was recently revealed as a potent (20 nM) ATP competitive Clk1 inhibitor with good selectivity over Clk3 via a unique binding mode. We have previously described a series of substituted 6-arylquinazolin-4-amines including NCGC00010037 (4, ML106) as potent inhibitors of Clk1 and Clk4 (Mott 2009).
Properties

ML196
NCGC00229610
| Physical & chemical properties | ||||
|---|---|---|---|---|
| Molecular Weight | 349.4 g/mol | |||
| Molecular Formula | C20H19N3O3 | |||
| cLogP | 3.5 | |||
| PSA | 65.5 Ų | |||
| Storage | ||||
| Solubility | ||||
| CAS Number | ||||
SMILES:
C1(NCC2COCC2)=NC=NC3=C1C=C(C4=CC5=C(OCO5)C=C4)C=C3
InChI:
1S/C20H19N3O3/c1-3-17-16(20(23-11-22-17)21-9-13-5-6-24-10-13)7-14(1)15-2-4-18-19(8-15)26-12-25-18/h1-4,7-8,11,13H,5-6,9-10,12H2,(H,21,22,23)
InChIKey:
ADJWQFUAOONPHD-UHFFFAOYSA-N
Activity
Summary activity statement /
The expansion of our previous efforts surrounding specific 6-arylquinazolin-4-amines as potent Clk and Dyrk1 inhibitors has yielded several agents with impressive potency and selectivity. ML197 (NCGC00185963, SID 85239750, CID 44223952); ML196 (NCGC00229610, SID 99380785, CID 46916178); and ML195 (NCGC00185981, SID 85239768, CID 44223970) are inhibitors that possess activity versus Clk1, Clk4 and Dyrk1A below 100 nM. A broad kinome scan has confirmed that these compounds are highly selective for these targets. Molecular docking studies highly suggest that they bind at the kinase hinge region. This work also defined the first reported inhibitor of Dyrk1B. These agents provide useful tool compounds to probe the role of these targets in pre-mRNA splicing and, in the case of Dyrk1B, specified roles in cancer.
In vitro activity - Selectivity assay
| Bioassay | (IC50) |
|---|---|
|
Clk1 (IC50) |
522 nM |
|
Clk2 (IC50) |
1055 nM |
|
Clk3 (IC50) |
3642 nM |
|
Clk4 (IC50) |
141 nM |
|
Dyrk1A (IC50) |
93 nM |
|
Dyrk1B (IC50) |
734 nM |
Summary /
ML196 (Compound 54) is found to be selective against the Clk4 and Dyk1A, compared to other Clks and Dyrk1 enzymes. IC50 values determined by Reaction Biology (www.reactionbiology.com). Compounds were tested in 10-dose IC50 mode with 3-fold serial dilution starting at 50 μM. Reactions were carried out at 10 μM ATP (Rosenthal, 2011).
In vitro activity - Molecular Modeling
Summary /
The homology model of Clk4 was developed by utilizing the X-ray structure of Clk1 as the template (86% sequence identity at the catalytic domain), while the Dyrk1B homology model was built based upon the highly homologous Dyrk1A (77% sequence identity) using MOE molecular modeling software (Figure 1A) (MOE). Several of our lead compounds were then docked into the ATP binding domains of these Clk and Dyrk1 models to achieve an optimal binding pose using FRED (OpenEye Scientific Software suite)(Figure 1B) (FRED). The resulting docking poses were considered in the context of the experimentally determined IC50 and Kd values. In agreement with our previous docking results, the quinazoline core adopted a common pose within the ATP binding pocket forming previously validated hydrogen bonds with the hinge region (Figure 1B highlights the docking of 46 with Clk4). As previously discussed, when an alkyl group was added to the 4-position amine (either a methyl or ethyl) activity generally improved. Our model rationalizes this result due to a small hydrophobic pocket (as indicated by a white line) in which the alkyl group is oriented which would likely increase specified van der Waals interactions and lock the inhibitor in a preferred conformation (Figure 1C). Interestingly, the SAR surrounding the amine side-chain suggests that several variations are well tolerated. This model suggests that the primary role of this moiety is space-filling rather than interacting with specific protein residues via H-bonding or electrostatic interactions. A primary hope was that these models might yield insight into defining more selective inhibitors of the Clk and Dyrk1 targets. One insight surrounds Clk4, which is nearly identical to Clk1 at the catalytic domain, but shows a key divergence in residue Asp248. Models docking 63 at Clk4 and Clk1 suggest that this molecule’s hydroxyl group is within an appropriate proximity to Asp248 (~3.2 Å) to form a hydrogen bond (Figure 1D). This potential interaction would help rationalize the potency for 63 versus Clk4 (136 nM) as compared to Clk1 (1522 nM). In the Dyrk1A crystal structure and the Dyrk1B homology model, the Asp residue at the same position shifted more than 6 Å away from the hydroxyl group in analogue 63 due to a backbone conformational change. This distance is too far for a viable hydrogen bond and offers a basis for the potency decrease versus Dyrk1A and Dyrk1B (>10,000 nM and 4420 nM, respectively). This interaction also highlights a potential route to furthering the SAR of this chemotype.
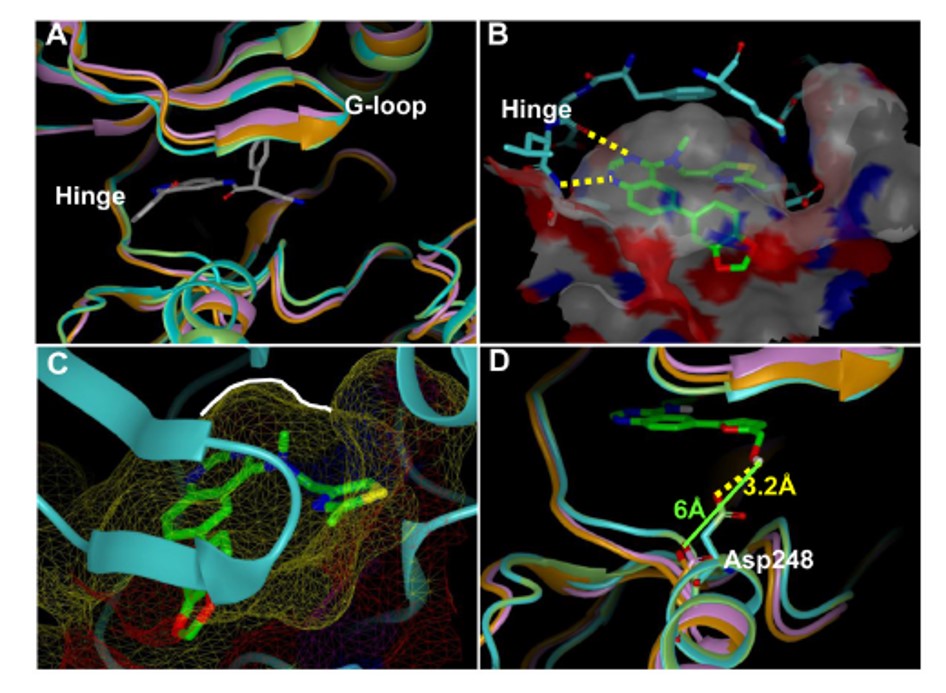
Figure 1. (A) Ribbon representation of the catalytic clefts in the Clk1 crystal structure (green; PDB 1Z57), Clk4 homology model (cyan), Dyrk1a crystal structure (orange, PDB 2VX3) and Dyrk1b homology model (purple). The ligand shown is the co-crystal ligand in Dyrk1A. (B) Docking model of 46 in the Clk4 catalytic cleft. The binding pocket is depicted by molecular surface and the hydrogen bonds are labeled as yellow dotted lines. (C) The close view of the small hydrophobic pocket (as indicated by a white line) in which the methyl group is sitting in the docking model of 46 within the Clk4 catalytic cleft. (D) Docking model of 63 in the Clk4 catalytic cleft superimposed with Clk1, Dyrk1a and Dyrk1b. The hydrogen bond from the hydroxyl group to Asp248 is labeled as a yellow dotted line. Clk4 kinase domain is a homology model derived from the X-ray structure of Clk1 (PDB code: 1Z57). This figure was prepared with the program VIDA (OpenEye Scientific Software).
In vitro activity - Kinase Proflie Dendogram
Summary /
To assure that these analogues still possessed the high selectivity for the Clk and Dyrk1 class of kinases we selected four representative inhibitors 45 (ML197), 46 (ML195), 54 (ML196), and compound 63 to examine in the same kinome-wide assay (possessing 442 kinases at the time of the profile). The kinome scan records activity as a percentage of kinase bound to an immobilized ligand in the presence and absence of each compound. In accordance with our previous work, activities beyond a selected threshold were submitted for Kd determination. The resulting Kd values further validated the selectivity of 45 and 54 for the Clk and Dyrk classes of kinases (Figure 3). Compound 46, although slightly less selective, is highly active against the desired targets as well as undesired kinases, Mek5 (Kd = 47 nM), a potential prostate cancer target41, and the kinase encoded by PIK3C2G (PI3K family)(Kd = 40 nM), which is involved in the pathophysiology of diabetes.42 The results for 63 suggested that this agent is somewhat promiscuous across several kinases and not acceptable as a probe of Clk and Dyrk1 activity (and highlights the utility of these profiles).
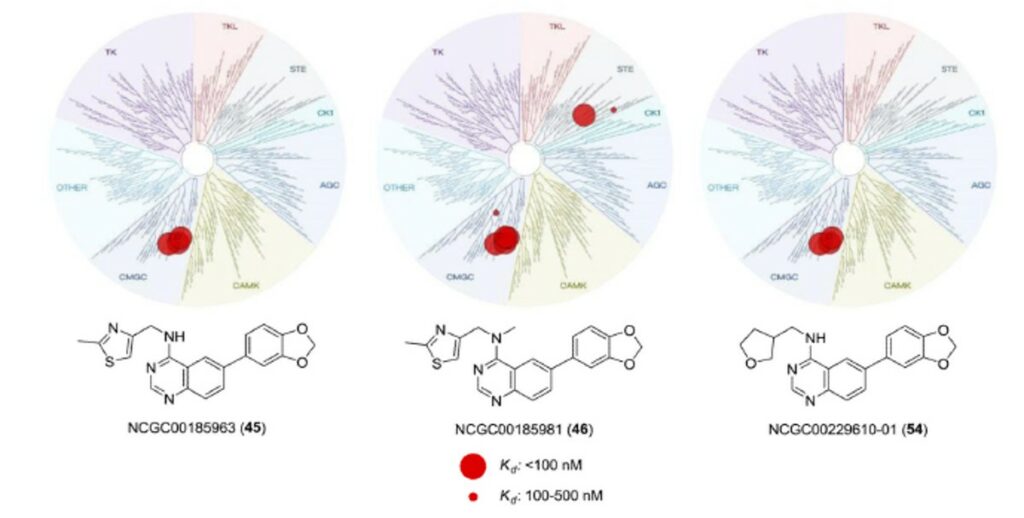
Figure 2. Dendrogram representation of the human kinome demonstrating kinase selectivity of reported inhibitors over a panel of 442 kinases. Activity for 45: Clk1 = 50 nM, Clk2 = 380 nM, Clk4 = 43 nM, Dyrk1A = 82 nM, PIP5K2C protein = 280 nM. Activity for 46: Clk1 = 18 nM, Clk2 = 59 nM, Clk4 = 5 nM, Dyrk1A = 13 nM, Dyrk1B = 300 nM, Dyrk2 = 480 nM, Erk8 = 430 nM, Mek5 = 47 nM, PIK3C2B protein = 340 nM, PIK3C2G protein = 40 nM, PIK3CG protein = 370 nM, PIP5K2C protein = 360 nM, Ysk4 = 190 nM. Activity for 54: Clk1 = 72 nM, Clk2 = 320 nM, Clk4 = 30 nM, Dyrk1A = 27 nM, PIK3C2B protein = 410 nM, PIK4CB protein = 430 nM, PIP5K2C protein = 310 nM. Data from DiscoveRx (http://kinomescan.com).
In vitro activity - ADME Profile
Summary /
The kinetic aqueous solubility and Caco-2 permeability data for ML197 (compound 45), ML195 (compound 46), and ML196 (compound 54) were obtained via a commercial provider (Analiza, Inc.) and (Cyprotex PLC). We further examined the stability of these agents in aqueous buffered environment for 48 hours (results are based upon retention of UV/Vis signal and mass assessment within a standard HPLC gradient). The outcome of these studies is shown in Table 1. ML197 and ML195 were found to possess only modest aqueous solubility while ML196 was highly soluble. The Caco-2 profile suggested that each agent was capable of passive membrane permeability and had favorable efflux ratios. Finally, each agent was highly stable in an aqueous environment for up to 48 hours.
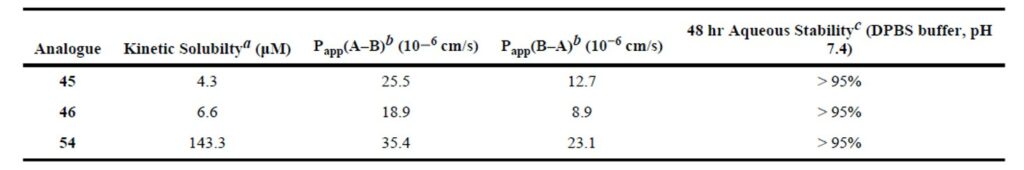
Table 1. Aqueous solubility and stability and Caco-2 permeability data for selected agents. (a) Solubility measurements done at Analiza, Inc (http://analiza.com). (b) Caco-2 permeability was measured at Cyprotex (www.cyprotex.com/adme). (c) Aqueous stability measured over 48 h in pH 7.4 DPBS buffer.
References
- qHTS for Inhibitors of CDC-like Kinase 4: Summary
- Rosenthal AS, Tanega C, Shen M, et al. Potent and selective small molecule inhibitors of specific isoforms of Cdc2-like kinases (Clk) and dual specificity tyrosine-phosphorylation-regulated kinases (Dyrk). Bioorg Med Chem Lett. 2011;21(10):3152-3158. doi:10.1016/j.bmcl.2011.02.114
- Prasad J, Manley JL. Regulation and substrate specificity of the SR protein kinase Clk/Sty. Mol Cell Biol. 2003;23(12):4139-4149. doi:10.1128/mcb.23.12.4139-4149.2003
- Black DL. Mechanisms of alternative pre-messenger RNA splicing. Annu Rev Biochem. 2003;72:291-336. doi:10.1146/annurev.biochem.72.121801.161720
- Wang Z, Burge CB. Splicing regulation: from a parts list of regulatory elements to an integrated splicing code. RNA. 2008;14(5):802-813. doi:10.1261/rna.876308
- Faustino NA, Cooper TA. Pre-mRNA splicing and human disease. Genes Dev. 2003;17(4):419-437. doi:10.1101/gad.1048803
- He C, Zhou F, Zuo Z, Cheng H, Zhou R. A global view of cancer-specific transcript variants by subtractive transcriptome-wide analysis. PLoS One. 2009;4(3):e4732. doi:10.1371/journal.pone.0004732
- Matlin AJ, Clark F, Smith CW. Understanding alternative splicing: towards a cellular code. Nat Rev Mol Cell Biol. 2005;6(5):386-398. doi:10.1038/nrm1645
- Jiang K, Patel NA, Watson JE, et al. Akt2 regulation of Cdc2-like kinases (Clk/Sty), serine/arginine-rich (SR) protein phosphorylation, and insulin-induced alternative splicing of PKCbetaII messenger ribonucleic acid. Endocrinology. 2009;150(5):2087-2097. doi:10.1210/en.2008-0818
- Schwertz H, Tolley ND, Foulks JM, et al. Signal-dependent splicing of tissue factor pre-mRNA modulates the thrombogenicity of human platelets. J Exp Med. 2006;203(11):2433-2440. doi:10.1084/jem.20061302
- Muraki M, Ohkawara B, Hosoya T, et al. Manipulation of alternative splicing by a newly developed inhibitor of Clks. J Biol Chem. 2004;279(23):24246-24254. doi:10.1074/jbc.M314298200
- Yomoda J, Muraki M, Kataoka N, et al. Combination of Clk family kinase and SRp75 modulates alternative splicing of Adenovirus E1A. Genes Cells. 2008;13(3):233-244. doi:10.1111/j.1365-2443.2008.01163.x
- Hartmann AM, Rujescu D, Giannakouros T, et al. Regulation of alternative splicing of human tau exon 10 by phosphorylation of splicing factors. Mol Cell Neurosci. 2001;18(1):80-90. doi:10.1006/mcne.2001.1000
- Perni, RB.; Bemis, J.; Nunes, JJ.; Szczepankiewicz, BG. Inhibitors of the CDC2-like Kinases and Methods of Use Therof. WO 2009/085226 A2. July 9. 2009
- Fedorov O, Huber K, Eisenreich A, et al. Specific CLK inhibitors from a novel chemotype for regulation of alternative splicing. Chem Biol. 2011;18(1):67-76. doi:10.1016/j.chembiol.2010.11.009
- Mott BT, Tanega C, Shen M, et al. Evaluation of substituted 6-arylquinazolin-4-amines as potent and selective inhibitors of cdc2-like kinases (Clk). Bioorg Med Chem Lett. 2009;19(23):6700-6705. doi:10.1016/j.bmcl.2009.09.121
- MOE Molecular Operating EnVironment. Vol. 2008.10. Chemical Computing Group Inc; Montreal, Canada: 2008
- FRED. OpenEye Scientific Software, Inc; Santa Fe, NM: 2010. http://www.eyesopen.com/


