ML216 : BLM (Bloom Syndrome Protein) Inhibitor
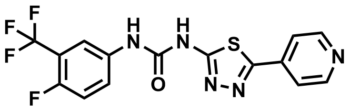
ML216
Target Name
Bloom Syndrome Protein
Target Alias
BLM
Target Class
DNA Helicase
Mechanism of Action
Inhibitor of BLM
Biological / Disease Relevance
Bloom’s Syndrome, DNA Repair Mechanism
In vitro activity
BLM helicase (DNA unwinding radio-labeled) assay (IC50)In vitro activity
BLM helicase (DNA unwinding fluorescent) assay (IC50)Target Information
BLM helicase is a DNA unwinding enzyme critical in DNA repair via the homologous recombination (HR) pathway. Mutations of the BLM gene result in diminished BLM helicase activity and can cause Bloom’s Syndrome, which is characterized by a long list of phenotypes, including predisposition to cancers. Similar to other DNA repair enzymes, inhibition of BLM helicase results in sensitization of tumor cells to conventional cancer therapies, such as camptothecin. Currently, there are no known small molecule inhibitors of BLM helicase; thus, the discovery of a novel small molecule inhibitor would help define the mechanism of action of the enzyme and provide a basis for future development of inhibitors and cancer therapeutics. ML216 displays low micromolar potency and selectivity over related helicases, such as RECQ1, RECQ5, and E. coli UvrD helicases. This probe also inhibits cell proliferation of BLM-proficient fibroblast cells (PSNF5) while having only minimal effects on BLM deficient fibroblast cells (PSNG13), indicating on-target activity in a cellular context. In addition, the probe molecule increases the frequency of sister chromatid exchanges, a diagnostic cellular phenotype consistent with the absence of a functional BLM protein.
Properties

ML216
NCGC00189393
| Physical & chemical properties | ||||
|---|---|---|---|---|
| Molecular Weight | 383.3 g/mol | |||
| Molecular Formula | C15H9F4N5OS | |||
| cLogP | 2.8 | |||
| PSA | 108 Ų | |||
| Storage | ||||
| Solubility | ||||
| CAS Number | 1430213-30-1 | |||
SMILES:
FC1=C(C(F)(F)F)C=C(C=C1)NC(NC2=NN=C(C3=CC=NC=C3)S2)=O
InChI:
1S/C15H9F4N5OS/c16-11-2-1-9(7-10(11)15(17,18)19)21-13(25)22-14-24-23-12(26-14)8-3-5-20-6-4-8/h1-7H,(H2,21,22,24,25)
InChIKey:
WMCOYUSJXXCHFH-UHFFFAOYSA-N
Activity
Summary activity statement /
ML216 (SID 99455330; CID 53385590) is the first selective BLM Helicase small molecule probe that inhibits with low micromolar potency. Its lack of activity against related helicases RECQ1, RECQ5, and UvrD (in E. coli) makes ML216 ideally suitable for use in helicase research and the broader field of DNA repair research. The anti-proliferative activity of the probe in BLMproficient fibroblast cells (PSNF5) and its increase of frequency in sister chromatid exchanges enhances the utility of the probe in cellular studies. Lastly, ML216 is a suitable starting point for further mouse tumor xenograft models and for the development of potential small molecule cancer therapeutics.
In vitro activity - Selectivity and Cytotoxicity Assay
| Bioassay | ML216 (IC50) |
|---|---|
|
BLM helicase (DNA unwinding radio-labeled gel-based assay) |
1.8 uM |
|
RecQ1 helicase (Anti-Target) |
~50 uM |
|
RecQ5 helicase (Anti-Target) |
>50 uM |
|
UvrD helicase (Anti-target) |
>50 uM |
|
BLM helicase (DNA unwinding fluorescent gel based assay) |
1.21 uM |
Summary /
ML216 is found to have > 28 fold selective against the BLM helicase vs. other helicases. The compound probe also showed no apparent cytotoxicity.
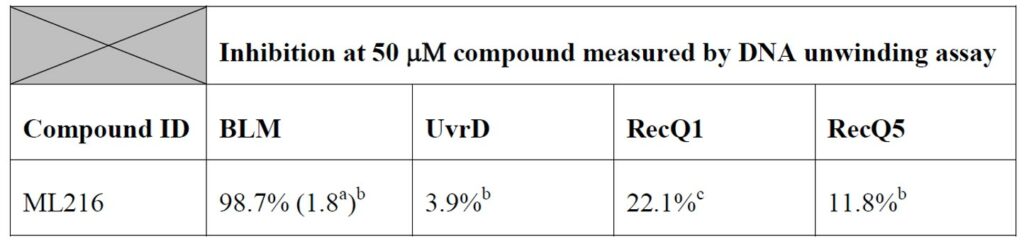
Table 1. Inhibition selectivity of ML216 on related helicases. (a) IC50 (μM) shown in parentheses, if applicable. (b) Determined by gel electrophoresis (32P labeled substrate). (c) Determined by helicase qHTS assay.
In vitro activity - BLM DNA unwinding assay
Summary /
The inhibition of DNA unwinding by both truncated and full-length BLM DNA unwinding for the probe ML216 is displayed in Figure 1.
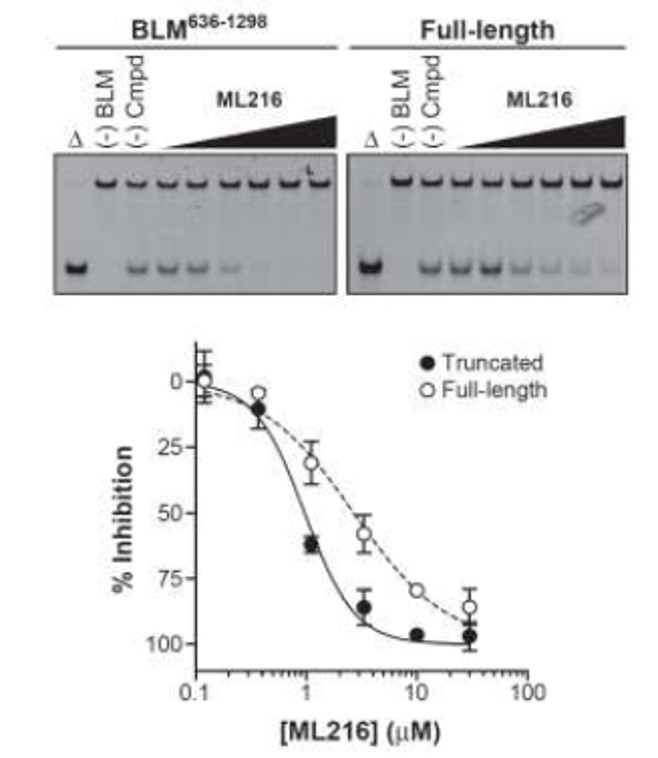
Figure 1. A) Representative gels for ML216 in the DNA unwinding gel electrophoresis assay using the truncated (left) and full-length (right) forms of BLM helicase and a fluorescent-labeled forked DNA substrate. B) Graphical representation of the percent inhibition of BLM by ML216. Each point represents the mean ± SD for three independent experiments.
Cellular activity - Cell proliferation assay: BLM-complemented (PSNF5) fibroblast
Summary /
ML216 selectively inhibits cell proliferation of BLM-proficient fibroblast cells (PSNF5) over BLM-deficient cells (PSNG13) (Figure 2), whereas the initial “hit molecule” 2 shows minimal effect on either cell line. More strikingly, ML216 caused a significant increase in the frequency of sister chromatid exchanges in PSNF5 cells, a key cytogenetic marker of cells lacking BLM activity (Figure 3). Together, these results serve to validate the ontarget cellular activity of ML216, as well as highlight a major advantage of the probe molecule over the initial hit.
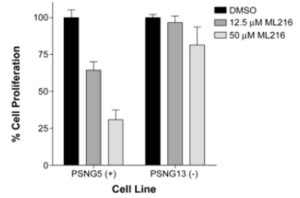
Figure 2. Inhibition of cell proliferation of BLM-complemented (PSNF5) fibroblast cell line, but not the BLM-deficient (PSNG13) fibroblast cell line after 72 hr treatment with ML216.
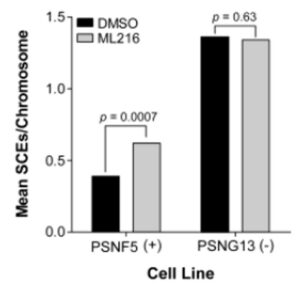
Figure 3. Increase in sister chromatid exchanges (SCEs) in the BLM-complemented (PSNF5) fibroblast cell line, but not in the BLM-deficient (PSNG13) fibroblast cell line after treatment with ML216 (50 μM).
In vitro activity - ADME profiling
Summary /
ML216 exhibits excellent mouse liver microsomal (MLM) stability and plasma stability (Table 2), indicating that the probe could prove to be useful in vivo. However, for this compound to have appropriate oral bioavailability, additional improvement in the aqueous solubility needs to be made.
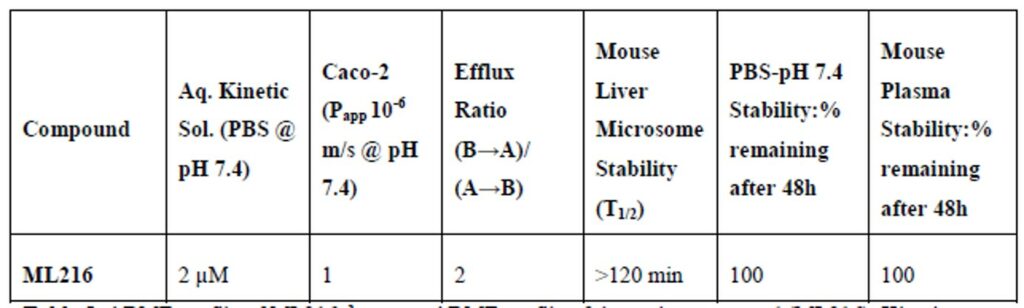
Table 2. ADME profile of ML216. (a) In vitro ADME profile of the probe compound (ML216). Kinetic solubility, Caco-2 permeability, MLM stability and plasma stability were all carried out by Pharmaron Inc. PBS buffer (pH 7.4) stability was performed in-house using LC/MS monitoring UV @ 254 and 220 nm.
In vitro activity - Mechanism of Action
Summary /
Besides the forked DNA duplex substrate, BLM has been shown to display an ATPdependent 3’-5’ DNA helicase activity that unwinds a variety of DNA substrates arising during DNA replication and repair such as Holliday junctions, G-quadruplex DNA, and displacement loops (D-loops). The BLM-dependent unwinding of such structures is also inhibited by ML216 (Figure 4), albeit to a lesser extent than the forked DNA duplex substrate. In addition, a fluorescence polarization assay, which monitored the binding of BLM to a fluorescently-labeled singled-stranded DNA oligonucleotide, was utilized to gain insight into the mechanism of action of ML216. As Figure 5 illustrates, ML216 inhibited the DNA binding of BLM with an IC50 of 5.1 μM, while having only a minimal effect on RecQ1 (IC50 = 101.8 μM). These results suggest the DNA binding domain of BLM as the putative binding pocket for ML216.
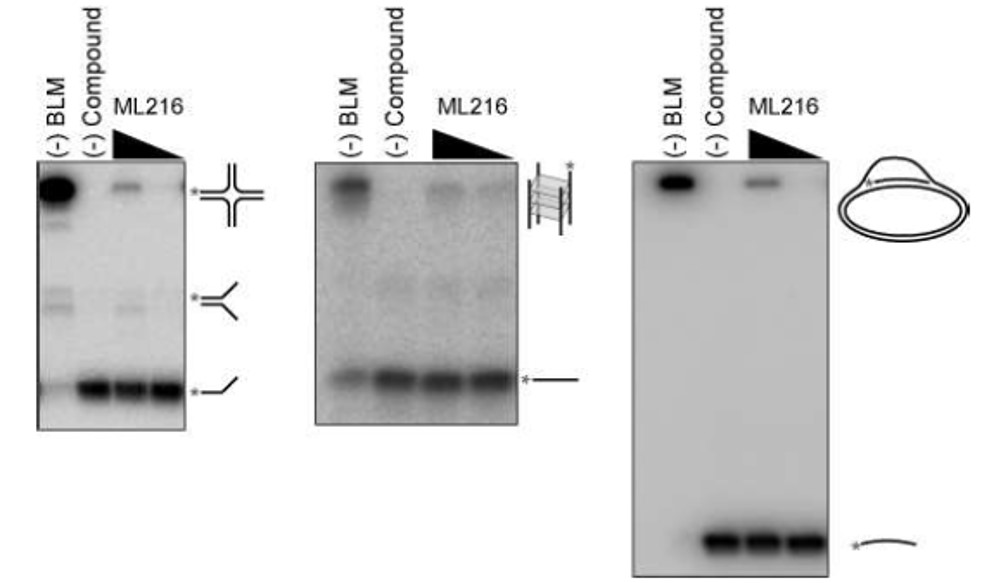
Figure 4. Inhibition of the processing of alternative DNA structures by BLM. Representative nondenaturing polyacrylamide gels showing the inhibition of BLM processing of Holliday-like junctions (left), G-quadruplex DNA (middle), and D-loop-like structures (right) by ML216 at 10 and 50 μM. Asterisks indicate location of 32P-radiolabel.
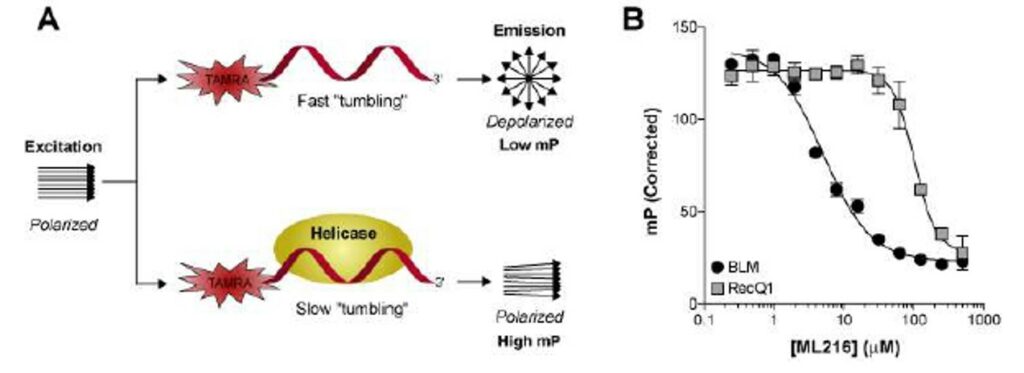
Figure 5. Inhibition of DNA binding by fluorescence polarization. A) Schematic diagram of the fluorescence polarization-based helicase-DNA binding assay. B) Inhibition of BLM and RecQ1 DNA binding by ML216.
References
- Probe Development Summary for Inhibitors of Bloom's syndrome helicase (BLM)
- Rosenthal AS, Dexheimer TS, Nguyen G, et al. Discovery of ML216, a Small Molecule Inhibitor of Bloom (BLM) Helicase. In: Probe Reports from the NIH Molecular Libraries Program. Bethesda (MD): National Center for Biotechnology Information (US); April 15, 2011


